Using R
Bio300B Lecture 2
Institutt for biovitenskap, UiB
25 August 2025
Basics of R
Why code?
Why not just use Excel for everything?
R as a calculator
Assigning
Assign object to a name
Forgetting to assign is a very common error
Functions
Function name followed by brackets
Arguments separated by comma
Don’t include an argument - uses default
Don’t need to name arguments if in correct order
Data types
Vectors
All elements must be the same type
Atomic vectors
Coercion
Automatic coercion
Predict the outcome of
[1] 1 0Subsetting a vector
Extract from
- first element
- last element
- second and third element
- everything but the second and third element
- element with a value less than 5
Matricies
2 dimensional
All elements same type
Arrays can have 3+ dimensions
Subsetting a matrix
[row_indices, column_indices]
[1] 4 5Lists
Each element of a list can be a different type
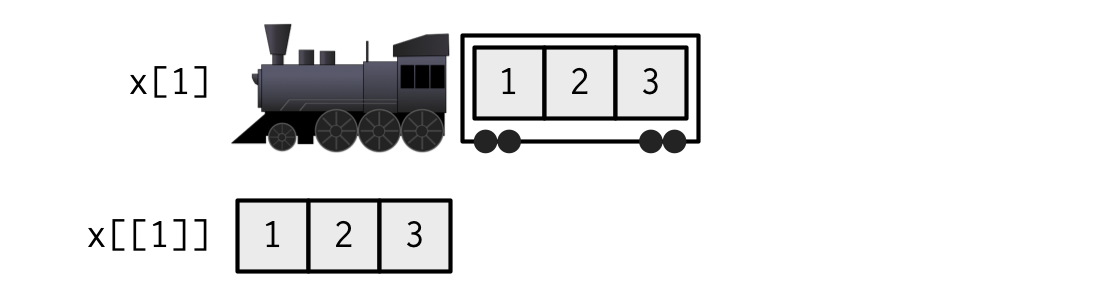
Subsetting a list
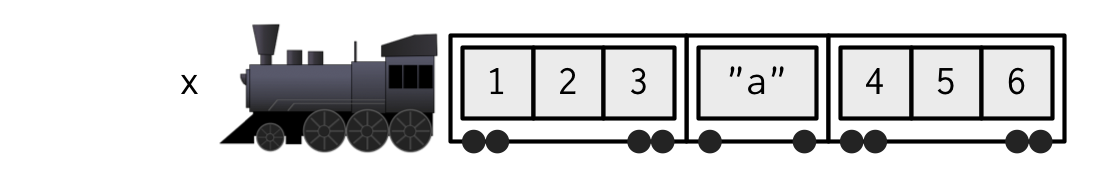
Can make a smaller list, or extract contents of a carriage
Named lists
Extract vector “a”
From the following extract
- The contents of the third element
- The third element still as a list
- The first and second elements as a list
Data frames and tibbles
rectangular data structure - 2-dimensions
columns can have different type of object
special type of list where all vectors have same length
Tibbles are better behaved version of data.frame
Data.frames have row and column names
Subsetting a tibble
With square brackets
With column names
Which method is safer?
Can also use dplyr package.
Extract the from
- the top ten rows
- the weight and Time columns
Logical tests
Return TRUE or FALSE
a == bTRUE if a equals b (test of equality - single=assignment)a != bTRUE if a not equal to ba > bTRUE if a greater than ba <= bTRUE if a less than or equal to b
Useful with subsetting, ifelse() or dplyr::case_when()
Boolean logic
logical conditions can be combined
&AND - TRUE if both TRUE|OR - TRUE if either TRUE!NOT - TRUE if FALSE
Control flow
if statements for choice
else is optional
Use && and || to return a single TRUE/FALSE
loops
Often don’t need an explicit loop - R is vectorised
for loops
for loops iterate over elements of a vector
for pitfalls
Need to pre-allocate space or slow
Rarely need a loop - purrr::map(), apply() generally cleaner
Better iteration with map()
$a
[1] 2
$b
[1] 5.5
$c
[1] 7 a b c
2.0 5.5 7.0 apply() for iterating over rows/columns of a matrix
Style
Code is communication
With your computer
With your collaborators
“Your closest collaborator is you six months ago but you don’t reply to email.” — Paul Wilson
- With reviewers/examiners
Need understandable code
Badstylemakescodehardertoread
Journal code archiving requirements
Nature Journals
A condition of publication in a Nature Portfolio journal is that authors are required to make materials, data, code, and associated protocols promptly available to readers without undue qualifications.
Canadian Journal of Fisheries and Aquatic Sciences
it is a condition for publication of accepted manuscripts at CJFAS that authors make publicly available all data and code needed to reproduce those results (including code to reproduce statistical results, simulation results, and figures) via an online data repository.
Tidy code
The only way to write good code is to write tons of shitty code first. Feeling shame about bad code stops you from getting to good code
— Hadley Wickham (@hadleywickham) 17 April 2015
- Makes code easier to read
- Makes code easier to debug
Make your own style - but be consistent
Naming Things
“There are only two hard things in Computer Science: cache invalidation and naming things.”
— Phil Karlton
- Names can contain letters, numbers, “_” and “.”
- Names must begin with a letter or “.”
- Avoid using names of existing functions - confusing
- Make names concise yet meaningful
- Reserved words include
TRUE,for,if
Which of these are valid names in R
cnames_1 = data_1
.columnNames()
.filter((c) => !['Timestamp', 'Score'].includes(c))
.map(d => d.replace(/^.*\[/, '').replace(/]$/, ''));
foldData_1 = data_1
.fold(aq.not('Timestamp', 'Score'), { as: ['question', 'answer'] })
.spread({question: d => op.split(d.question, '[')} , {as: ['question_title', 'question']})
.derive({question: d => op.replace(d.question, ']', '')});Plot.plot({
marginLeft: 170,
marginBottom: 60,
height: 500,
x: {label: 'Frequency', labelOffset: 50},
y: {label: null, domain: cnames_1},
color: {
legend: true,
domain: ["Yes", "No"],
range: ["green", "orange"],
},
style: {fontSize: '25px'},
marks: [
Plot.barX(foldData_1, {y: 'question', x: 1, inset: 0.5, fill: 'answer', sort: 'answer'}),
Plot.ruleX([0])
]
})Names can be too long
Or too short
k
Naming convensions
| camelCase 🐫 | UpperCamelCase | snake_case 🐍 |
|---|---|---|
| billLengthMM | BillLengthMM | bill_length_mm |
| bergenWeather2022 | BergenWeather2022 | bergen_weather_2022 |
| dryMassG | DryMassG | dry_mass_g |
| makeWeatherPlot | MakeWeatherPlot | make_weather_plot |
White-space is free!
Place spaces
- around infix operators (
|>,+,-,<-, ) - around
=in function calls - after commas not before
Good
Bad
Split long commands over multiple lines
Indentation makes code readable
Good
Bad
Stylers & lintr
Use styler package to edit code to meet style guide.
Use lintr package for static code analysis, including style check
Comments for navigation
Helps you find your way around a script
No magic numbers
Split analyses over multiple files
Long scripts become difficult to navigate
Fix by moving parts of the code into different files
For example:
- data import code to “loadData.R”
- functions to “functions.R”
Import with
Number files so they sort alphabetically in order of use.
Don’t repeat yourself
Repeated code is hard to maintain
Make repeated code into functions.
Single place to maintain
Comments
Use # to start comments.
Help you and others to understand what you did
Comments should explain the why, not the what.
Try to make code self-documenting with descriptive object names