Enough targets to Write a Thesis
For this tutorial you will need:
- RStudio (version 2022.2 or newer)
- R
- quarto
- quarto R package (
install.packages("quarto")) - targets R package (
install.packages("targets")) - tarchetypes R package (
install.packages("tarchetypes"))
If quarto is new to you, have a look at the Reproducible documents tutorial (REF to Quarto book) before proceeding here.
1 Introduction
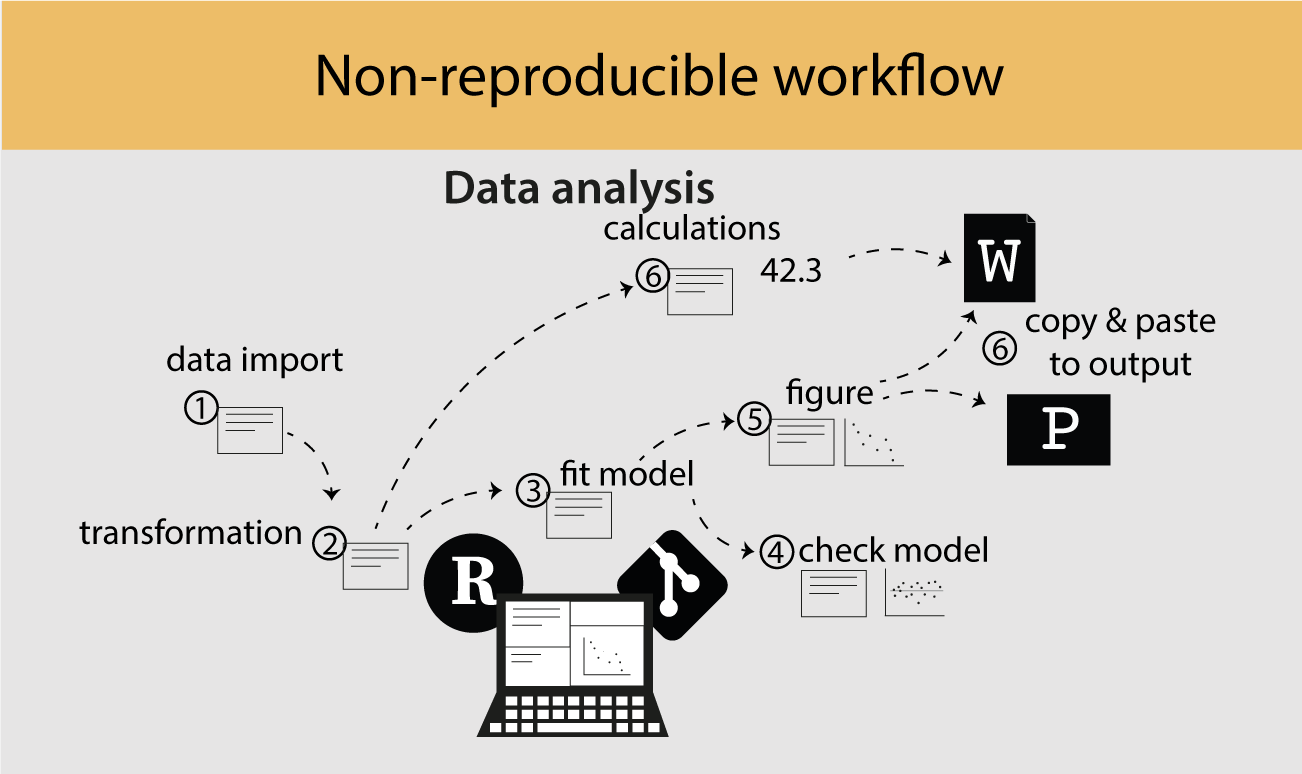
Writing a thesis requires importing data (1), writing code to clean, transform (2), analyse data (3), and making figures (5) and writing (6; Figure 1.1). This is done by writing several R scripts and running one script after another producing results and figures. All the time, the code is updated, to add new data, transforming the data, changing analysis or adding a figure. Often many iterations of running the same scripts are needed and it is difficult to keep track of which scripts need rerunning. In addition, complex and computational heavy data analysis can take a lot of time especially when it requires rerunning the analysis.
This workflow is very inefficient, non-reproducible and error prone. There is a better way and this tutorial will show you how.

In this chapter we will first explain the concept of abstraction and then go through the basic workflow of a targets pipeline. We will use plant trait data from two sites on Svalbard. One site is located close to nesting sites of sea birds receiving nutrient input while the other site is a reference location and has minimal nutrient input by sea birds.
Follow the instructions below to download the repo containing data and code.
To download the R project, run:
# install.packages("usethis") # if you don't have it already.
usethis::use_course("biostats-r/targets_workflow_svalbard")Then follow the instructions. This will open the targets-workflow-svalbard Rstudio project.
