13 Tidying data with tidyr
13.1 Tidy data
Before you can start analysing or plotting data, you often need to tidy it. Tidy data is a standardised way to structure a dataset which makes it much easier to process, analyse and plot the data. Functions in the tidyr and dplyr packages, both part of tidyverse, can be very useful for tidying data.
Tidy data is a standard, consistent way to organize tabular data. Briefly, tidy data follows a short series of rules:
- each variable in the data set is presented in a specific column,
- each observation in the data set is presented in a specific row,
- each cell at the intersection of a row and a column contains a single value.
The following figure illustrates these rules.

The dataset presented in Table 11.1 respects all three rules, and is thus a tidy dataset.
On the contrary, the following dataset is not tidy:
| Date | 17.06.2016 | 21.06.2016 | 23.06.2016 | 26.06.2016 | 27.06.2016 | 28.06.2016 | 29.06.2016 | 30.06.2016 | 02.07.2016 | 04.07.2016 | 05.07.2016 | 06.07.2016 | 08.07.2016 | 10.07.2016 | 11.07.2016 | 13.07.2016 | 15.07.2016 | 17.07.2016 | 19.07.2016 | 21.07.2016 | 23.07.2016 | 24.07.2016 | 26.07.2016 | 28.07.2016 | 30.07.2016 | 02.08.2016 | 04.08.2016 | 06.08.2016 | 11.08.2016 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time | 10:00 | 11:00 | 11:00 | 13:00 | 13:00 | 09:33 | 13:00 | 15:00 | 09:00 | 09:15 | 09:30 | 09:15 | 09:15 | 09:15 | 09:15 | 09:30 | 10:00 | 13:00 | 17:30 | 10 | 10:00 | 09:30 | 09:30 | 09:30 | 09:15 | 09:30 | 15:00 | 11:00 | 15:00 |
| Weather | Sunny_PartlyCloudy | Cloudy_Fog | Cloudy_Sunny | Cloudy_Rainy_Windy_Fog | Cloudy_Windy_Rainy | Windy_Rainy_MostlyCloudy | Windy_Sunny_Cloudy | Windy_Cloudy | Windy_Cloudy | Cloudy_Sunny | Cloudy_Sunny | Cloudy_Fog_Rainy | Sunny_PartlyCloudy | Cloudy_Rainy | Cloudy | Cloudy_Sunny | Cloudy | Cloudy_Rainy | Cloudy_Rainy | Cloudy_Sunny | Sunny_Cloudy | Cloudy_Sunny | Cloudy_Rainy | Cloudy_Rainy | Cloudy_Rainy | Cloudy_Rainy | Cloudy_Rainy | Cloudy_Rainyn | Cloudy_Snowy_Sunny |
| Observer | LV_AH | LV | LV | LV | LV_SB | LV_SB | LV_SB | LV | LV_SB | SB_LV | SB_LV | LV_SB | LV_SB | LV_SB | LV | LV | LV_SB_AH | SB_LV | SB_LV | SB_LV | SB_LV | LV | LV | LV | LV | LV | LV | LV | LV |
| E01a | 0 | NA | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 2 | 3 | 3 | 6 | 8 | 8 | 11 | 13 | 10 | 9 | 7 | 3 | 6 | 3 | 3 | 1 | 1 | 1 | 2 | 1 |
| E01b | 0 | NA | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 3 | 3 | 8 | 19 | 23 | 26 | 24 | 16 | 16 | 8 | 5 | 2 | 3 | 4 | 3 | 2 |
| E01c | 0 | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 5 | 9 | 12 | 10 | 7 | 4 | 3 | 3 | 3 | 2 | 2 | 0 | 1 | 1 |
| E01d | 0 | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 2 | 4 | 3 | 2 | 4 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 0 |
| E01e | 0 | NA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 4 | 4 | 5 | 4 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
Indeed, at least one of the rules is broken since columns display data matching several variables (Date, time, Weather, etc).
Importing data from a file containing tidy data is a great way to start your work, but it is not a prerequisite to data import. As long as your data is tabular, you will be able to import it in R, and tidy it later.
The process of tidying data is to convert the data you have to data that meets this standard.
13.2 Multiple values in a cell
If a cell contains multiple values, we need to separate to tidy the data. We can separate the values into multiple columns with tidyr::separate_wider_delim(). For example, in this small dataset, site code and plot number have been combined into one column separated by a hyphen.
dat <- tribble(
~id, ~value,
"A-1", 1,
"A-2", 2,
"B-1", 3
)
dat# A tibble: 3 × 2
id value
<chr> <dbl>
1 A-1 1
2 A-2 2
3 B-1 3We can use separate_wider_delim() to split site and plot into separate columns. The delimited is set by the delim argument.
dat |>
separate_wider_delim(cols = id, delim = "-", names = c("site", "plot"))# A tibble: 3 × 3
site plot value
<chr> <chr> <dbl>
1 A 1 1
2 A 2 2
3 B 1 3Related function, separate_wider_regex(), which uses regular expressions to split the column, can be more flexible.
If there are multiple measurements of the same type in a single cell, then tidyr::separate_longer_delim() can be useful to separate the data into multiple rows.
13.3 Reshaping data - wide to long
It is very common to need to reshape data to make it tidy. This can be done with the pivot_* functions.
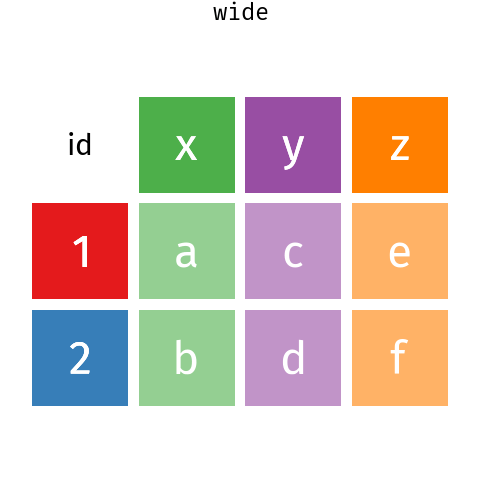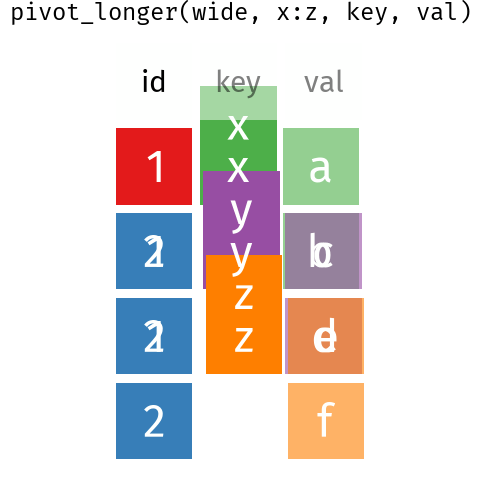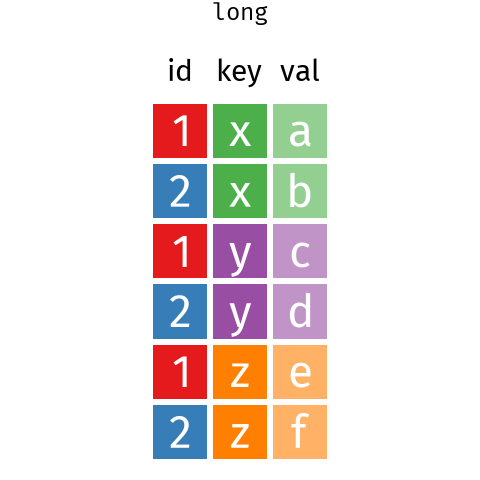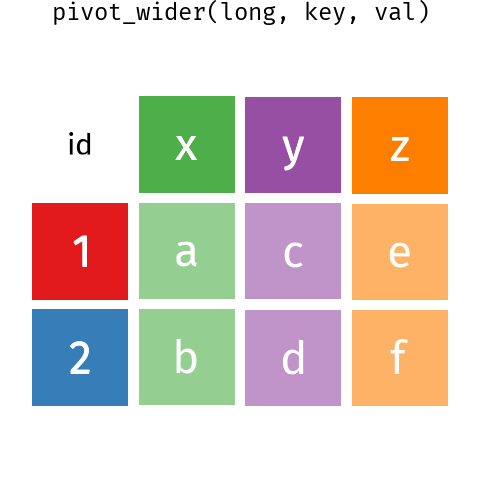
pivot_longer() and pivot_wider() .These are some Bergen climate data from Wikipedia (NB for demonstration only wikipedia is not a good source of climate data - use seklima for Norwegian data).
Rows: 4 Columns: 13
── Column specification ────────────────────────────────────────────────────────
Delimiter: "\t"
chr (1): Måned
dbl (12): Jan, Feb, Mar, Apr, Mai, Jun, Jul, Aug, Sep, Okt, Nov, Des
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.# A tibble: 4 × 13
Måned Jan Feb Mar Apr Mai Jun Jul Aug Sep Okt Nov Des
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Norma… 3.6 4 5.9 9.1 14 16.8 17.6 17.4 14.2 11.2 6.9 4.7
2 Døgnm… 1.7 1.7 3.3 5.8 10.4 13.1 14.2 14.2 11.5 8.8 4.8 2.7
3 Norma… -0.4 -0.5 0.9 3 7.2 10.2 11.5 11.6 9.1 6.6 2.8 0.6
4 Nedbø… 190 152 170 114 106 132 148 190 283 271 259 235 This might be a nice way to present the data but it is not tidy data: each row is not an observation; each column is not a variable.
We can use pivot_longer() to reshape the data. The months, selected by the cols argument in the column names (see Section 14.2.1 for more on the syntax used here) will become a new variable with a name set by the names_to, and the data values get put into a column named by the values_to argument.
bergen_klima_long <- bergen_klima |>
pivot_longer(cols = Jan:Des, names_to = "Month", values_to = "value")
bergen_klima_long# A tibble: 48 × 3
Måned Month value
<chr> <chr> <dbl>
1 Normal maks. temp. °C Jan 3.6
2 Normal maks. temp. °C Feb 4
3 Normal maks. temp. °C Mar 5.9
# ℹ 45 more rowsThe data are now tidier, but it would probably be more useful to reshape the data again, and have a column for each climate variable. We can do this pivot_wider().
13.4 Reshaping data - long to wide
We can tell pivot_wider() which column contains what will become the column names and the data with the names_from and values_from, respectively.
bergen_klima_wider <- bergen_klima_long |>
pivot_wider(names_from = "Måned", values_from = "value")
bergen_klima_wider# A tibble: 12 × 5
Month `Normal maks. temp. °C` `Døgnmiddeltemp. °C` `Normal min. temp. °C`
<chr> <dbl> <dbl> <dbl>
1 Jan 3.6 1.7 -0.4
2 Feb 4 1.7 -0.5
3 Mar 5.9 3.3 0.9
4 Apr 9.1 5.8 3
5 Mai 14 10.4 7.2
6 Jun 16.8 13.1 10.2
7 Jul 17.6 14.2 11.5
8 Aug 17.4 14.2 11.6
9 Sep 14.2 11.5 9.1
10 Okt 11.2 8.8 6.6
11 Nov 6.9 4.8 2.8
12 Des 4.7 2.7 0.6
# ℹ 1 more variable: `Nedbør (mm)` <dbl>The data are now in a convenient format for plotting or analysis.
With the Mt Gonga biomass data downloaded previously, pivot the data so that the height data (H1-H10) are in one column.
Hint
pivot_longer
Contributors
- Richard Telford
